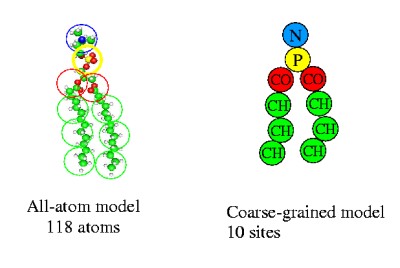

Solvent may be removed as well. Within such model, much bigger systems consisting of thousand of lipids can be simulated. The principal problem of such an approach is building the interaction potentials for the coarse-grained model. They may be constructed from all-atomic molecular dynamics simulations of just a few lipids using the inverse Monte Carlo technique as described in the paper: A.P.Lyubartsev, "Multiscale modeling of lipids and lipid bilayers", Eur. Biophys. J., v. 35, 53-61, 2005.
A movie of the process of self-assembly of 392 lipids, which were initially randomly dispersed in a periodic cubic box of 20 nm, can be downloaded and viewed on the link below. The simulation was performed using molecular dynamics with local Lowe-Andersen thermostat. Non-scaled time is shown.
Membrane self-assembly (.mpg file)
The data file containing the short-range part of the interaction potential for the coarse grained DMPC lipid model can be picked up here: dmpcCG.pot It contains values of potentials, in kJ/M, for all pairs of 4 interaction sites (N,P,CO and CH labelled as 1,2,3,4 correspondingly) as well as intramolecular bond potentials. The electrostatic interactions due to charges (+1) at N and (-1) at P should be added also.
An ESPRESSO script to run a coarse-grained simulation of lipids, together with the tabulated effective potentials, can be found here.