 |
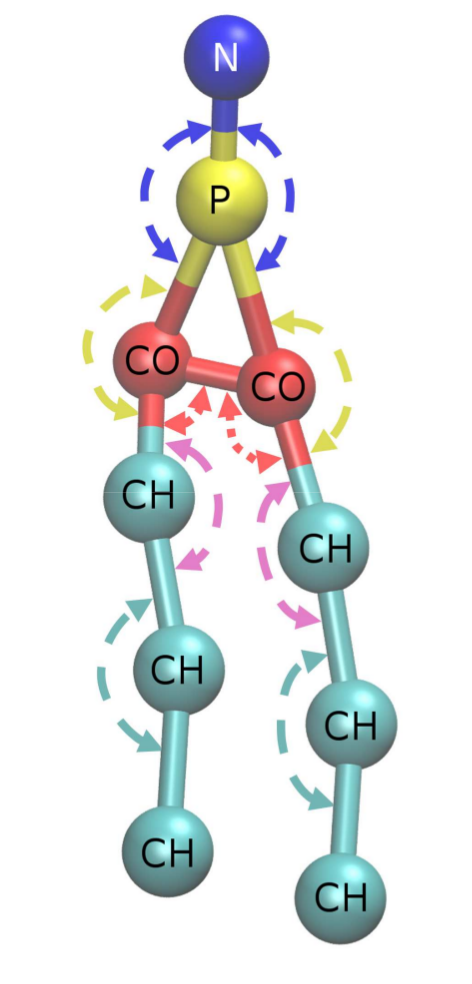
&Parameters TrajFile = cgtraj.dmpc16-400ns.xmol NMType = 1 NameMType = dmpc_NM.CG.mmol NMolMType = 16 OutputFile = dmpc16-100aa.rdf RMaxNB = 20. RMaxB =10.0 ResolNB =0.1 ResolB=0.02 ResolA=1.0 &ENDParameters &CGTypes N:N P:P CH:C2 C3 C4 C6 C7 C8 CO:C1 C5 &EndCGTypes &RDFsNB Add: all # Add: N--P # Add: N--P: N P # Add: N--CO: N C1, N C5 &EndRDFsNB &RDFsB add: dmpc_NM.CG: 1: N P add: dmpc_NM.CG: 2: P C1, P C5 add: dmpc_NM.CG: 3: C2 C3, C3 C4, C6 C7, C7 C8 add: dmpc_NM.CG: 4: C1 C2, C5 C6 add: dmpc_NM.CG: 5: C1 C5 &EndRDFsB &RDFsA Add: all #add: dmpc_NM.CG: All #Other way - generate all A-bonds for the molecule #add: dmpc_NM.CG: 6: N P C1 #Example - create N-P-CO A-bond &EndRDFsA
 |