


Next: .mmol
Up: File formats
Previous: File formats
Contents
.xmol
This is plain text single configuration or trajectory file format, which can be produced by many molecular
modeling packages, including MDynaMix, MagiC, or converted from *.gro by gro2xmol.py script gro2xmol.
It consists of a number of consequent frames, with each frame having the following structure:
- line 1:
- Number of atoms in the frame (N)
- line 2:
- A commentary line
- line 3:
- Name(atom1) X(atom1) Y(atom1) Z(atom1)
- line 4:
- Name(atom2) X(atom2) Y(atom2) Z(atom2)
- lines 5,6...,N+2:
- Names and coordinates of atoms 3 - N.
In case of trajectory, configuration files from each time frame are written consequently one after another.
There is no common requirements for the commentary (second) line. For CGtraj module of MagiC it is
assumed that the second line of each configuration follows this format (accepted in MDynaMix):
(char) <time> (char-s) BOX: <box_x> <box_y> <box_z>
where (char) is any character word, <time> is time in 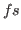 ,
, <box_x> <box_y> <box_z>
(following after keyword BOX). The length unit is Ångströms and time unit is femtoseconds.
The time step information is not needed for CGTraj but the box size information is essential.
If no box size information is present in the trajectory, the box size information can be supplied
in the input file, but the later has a sence only in constant-volume simulations.
In xmol-files produced by MagiC the second line may have no time-stamp and box sizes.



Next: .mmol
Up: File formats
Previous: File formats
Contents
Alexander Lyubartsev
2016-05-03