J.Phys.Chem. Vol. 101, p.4335 (1997)
Monte Carlo Simulation Study of DNA Polyelectrolyte Properties in the Presence of Multivalent Polyamine Ions.
Alexander P. Lyubartsev† and Lars Nordenskiöld*
Division of Physical Chemistry, Arrhenius Laboratory, Stockholm University
S 106 91 Stockholm Sweden.
Abstract
Properties of ionic distributions of mixtures of atomic (Na+, Mg2+, Cl--) and molecular (spermidine3+) ions in the vicinity of DNA have been studied by the Monte Carlo simulation method, within the frame of a continuum dielectric approach. Several models differing by the level of approximations have been considered and compared. In one of these models the DNA phosphate charge distribution is described by point charges in a double helical array corresponding to B-DNA, thus in an idealized way incorporating the major and minor groove structure. Ion distribution profiles, ion binding properties and competition effects in the association of the different counterions to DNA have been studied. Particular attention has been paid to the importance of the discrete distribution of the three charges on the spermidine molecular model. The work has shown that the behavior of complex multivalent ions differs strongly from the properties of simple metal ions of the same valence. The competitive behavior and binding properties of the spermidine molecule modeled with distributed charges, appears to be reduced from that expected for point charge (or spherical) three-valent ions, even compared to the predictions of the Poisson-Boltzmann approximation, and more likely correspond to the behavior of atomic divalent ions. The results of binding properties was found to be in qualitative agreement with existing NMR diffusion studies of polyamine association to DNA.
_________________________
† also affiliated with Institute of Physics, St.Petersburg State University, 198904, St.Petersburg, Russia. e-mail: sasha@tom.fos.su.se
* e-mail: lnor@tom.fos.su.se
1. Introduction
Many of the physical properties of DNA are strongly dependent on electrostatic interactions between the negatively charged DNA polyelectrolyte and the surrounding positively charged counterions1-3. In a large number of studies the polyelectrolyte properties of DNA have been studied and discussed in detail. The existing theoretical approaches include the counterion condensation theory of Manning1,4, the Poisson-Boltzmann (PB) (or self-consistent field) theory5,6, hypernetted-chain equations7,8 and computer simulations9-13. Most of the works deal with solutions of monovalent ions within the frame of the so-called primitive model which treats the ions as charged spheres and DNA as a hard uniformly charged cylinder, imbedded in a dielectric continuum described by a dielectric constant9-12. In a few studies the distributions of divalent ions around a cylindrical DNA model was investigated10,12-15. Furthermore, the problem of competitive binding between counterions of different valences has also been studied11,16,17. The discrete helical charge distribution of DNA has also been accounted for in some papers3,18-21. Various numerical studies over the past decades have shown that, among simple theories, the PB equation provides a more solid basis for studying polyelectrolytes than e.g. approaches based on Mannings condensation theory (see reviews22,23). The results of both computer simulations and hypernetted-chain theory have also shown that while for monovalent ions the Poisson-Boltzmann equation generally satisfactorily describes the properties of the DNA ionic environment, for multivalent ions it noticeably underestimates the ion density in the nearest vicinity of the polyion.
The interaction of DNA with complex multivalent positively charged molecular ions, such as e. g. the multivalent polyamine molecules ( like spermidine3+ and spermine4+), is of considerable interest due to the question of the biological function of the polyamines and their interactions with nucleic acids (paper24 and references cited therein). The problem of the distribution of these kind of multivalent counterions around DNA has been less studied theoretically despite their biological significance and numerous experimental studies24-31. In the crystal form, X-ray works25-27 have shown that polyamines bind in a specific manner to well-defined but varying sites on DNA. In contrast, DNA-polyamine interactions in solutions studied by NMR and other techniques clearly demonstrate association to DNA in a loose electrostatic, non-specific manner28-30 which can qualitatively be well described by polyelectrolyte theory.
The question of the interaction between the DNA polyelectrolyte and complex multivalent polyamines such as spermidine is interesting also from a theoretical point of view. The spermidine molecule combines properties of a three-valent point charge ion and of a monovalent ion. Thus, the spermidine density distribution outside a DNA polyelectrolyte, r, in an external field y, is at a long length scale (compared to the length of the polyamine molecule) approximately given by: r~ exp(-3ey/kT) in the low concentration limit, as for three-valent ions. This creates a high concentration of spermidine molecules around DNA even at very low bulk concentration. On the other hand, at small distances from the DNA surface, each of the three charged groups of a spermidine molecule may be considered as a monovalent ion. Clearly, the behavior of molecules with distributed charges is expected to be different from that of a "simple" (atomic) three-valent ion which have the charge at one point. A simple scaling and a mean field analysis of the influence of ligand spatial organization on electrostatic binding to DNA was given in a recent paper32. In another paper33, effects of ligand structure was taken into account using lattice statistics within the frame of a PB approach. In the present work we have applied the Metropolis Monte Carlo computer simulation method to study the ionic distributions and binding properties of the polyamine spermidine3+ with DNA. The systems that are modeled also contain simple mono- or divalent ions that are able to compete with spermidine for association to DNA. We consider and compare several models differing by the level of the approximations used. A particular issue that we address is the importance of the distribution of the three discrete charges on the spermidine molecule, for determining the association of this polyamine to DNA and the effect of competition with mono- and divalent counterions.
2. Models
A. Hard cylinder in the Poisson-Boltzmann approximation (PB model).
Our simplest approximation is the well-known Poisson-Boltzmann theory for ion distributions around a hard, uniformly charged cylinder5,6. The charge density of a-species of ions at the distance r from the polyion axis in the Poisson-Boltzmann (mean field) approximation is
![]()
where ca is the bulk concentration, y(r) =ef(r)/kT is the reduced electrostatic potential defined in the case of cylindrical symmetry by:
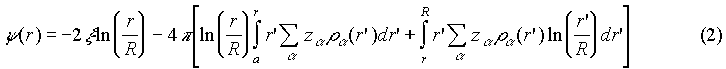
x is the reduced linear charge density of the polyion, a is the polyion radius (x=-4.2 and a=10Å for B-DNA), and R is the radius of the "cell model"34 defined by the polyion (phosphate) concentration cp:

(NA is the Avogadro number and lb=e2/(ekT) is the Bjerrum length).
In the Poisson-Boltzmann theory a specific character of an ion reveals itself only in the valence and, optionally, in the distance of minimum approach to the polyion. Equations (1) and (2) can easily be solved numerically and this approach has been widely used for studies of the ionic environment of DNA and for interpretation of various experimental data5,6,23,35,36.
In our following models we take into account some points neglected in the simple mean field theory. We use the Monte Carlo simulation technique to simulate ions around DNA in all of the models B,C and D below. By doing so we fully take into account the ion correlations neglected in the mean field theory (eq.(1)). The difference between the different Monte Carlo models lies in the level of detail in the description of the charge distribution of the DNA and spermidine molecules.
B. Spherical ions around a cylindrical DNA (SI model).
In this model we consider the ions as charged particles with a repulsive short-range potential in a dielectric medium of permitivity e=78. The spermidine molecules are also modeled as spherical ions. The ion-ion potential is given by:
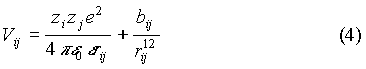
where bij=kT((si+sj)/2)12 and rij=|ri-rj|, si being the "hydrated" ion diameter. The comparison of the PB and SI models allows us to evaluate the importance of ion correlations on the properties of the ion distributions.
C. Chain model of spermidine, hard cylindrical model of DNA (HC model).
In this model we present an idealized spermidine (or methylspermidine) molecule as a chain of three monovalent ions linked by bonds with a harmonic potential:
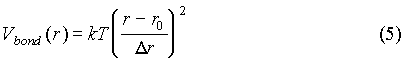
where r is the distance between the monomers, r0 is an equilibrium distance and Dr is a characteristic deviation. In our work we use values r0=5.6 Å and Dr=0.5 Å, chosen in order to model an idealized spermidine molecule. Each monomer of a spermidine molecule interacts with other monomers and with other ions by the potential in Eq. (4).
D. Grooved model of DNA (GD model).
This model19,20 is devised to incorporate effects of the helical DNA grooves and the discrete charge localization on the DNA surface. The DNA model has a hard cylindrical core of radius a=8 Å and charged "phosphate groups" situated at the sites corresponding to the B-form of DNA. Each phosphate group has a charge -e and a soft repulsive r-12 potential of effective diameter sp=4 Å, the phosphate - ion interactions also being described by the potential given by Eq. (4). This set of phosphate groups forms an idealized model of double-stranded DNA with two grooves. Due to the repulsive short-range interactions, the average effective radius of DNA is about 10 Å. The simple ions and the spermidine molecules are treated in the same way as in the HC model.
In all our models we consider the solvent as a dielectric medium. The ion-ion and ion-phosphate potentials in Eq. (4) represent an approximation to the solvent-mediated effective potential between the ions in the solution. This means that e in eq. (4) is a parameter of the effective ion-ion potential and not necessarily represents a local dielectric constant which, according to some estimations37, may assume much lower values in the first two solvent molecular layers next to a charged surface, as compared to the bulk water. Existing studies of molecular-ion systems at a molecular level, have shown that the effective (mean force) potentials of ions in water usually have one or two oscillations around the Coulomb potential with e=78 for small (few Ångströms) distances between the ions38-40. These oscillations reflect the molecular nature of the solvent. Still, for many applications, including strong polyelectrolytes and high salt concentrations, continuum dielectric models have been successful (see e.g. reviews38,41). Our choice of the continuum dielectric model has the intention to study purely electrostatic effects.
The ion diameter si has the sense of a hydrated ion diameter and is an adjustable parameter of the model that includes effects of the molecular nature of the solvent in an averaged form. The most suitable values of si for specific ions can be obtained e.g. from fitting to osmotic coefficient data (see refs.42,43). In this work we have used si=4Å which is typical for alkali ions and for charged aminogroups.
3. Simulation details
In all our simulations we use an hexagonal simulation cell with DNA located along the z-axis and with periodic boundary conditions in all directions. Thus we really simulate an ordered DNA system14,19. Our previous simulations19 showed that ion distribution results for the cylindrical cell model and the hexagonal models are indistinguishable at least for inter-axial DNA-DNA distances 2R larger than 100 Å. The long-range part of the electrostatic interaction was taken into account by the Ewald method. Details of application of the Ewald method to a hexagonal cell are given in our previous work19. The height of the simulation cell was in most cases 102 Å, corresponding to three full turns of B-DNA with 60 phosphate groups. The size of the cell in the perpendicular direction was determined by the DNA (phosphate) concentration and was typically about 100 Å. The total number of simulated ions varied from 100 to 1000 depending on salt concentration.
Most of the simulations were carried out within the NVT ensemble using the Monte Carlo method. The spermidine molecules were found to move very slowly by applying the standard Metropolis algorithm, slowing down the convergence of the calculations, particularly at low spermidine or DNA concentrations. In these simulations we have used 2.5.107 MC steps for equilibration of the system and the same number of steps for the average collection. Some simulations at low concentrations were carried out in the grand-canonical (mVT) ensemble. Performing the mVT Monte Carlo algorithm including also removal or installation of ions provides faster sampling of the configuration space and, consequently, faster convergence. This enabled the beginning of averaging already after 2.106 MC steps. On the other hand, in the grand canonical calculations for complex ion mixtures, it was difficult to guess the chemical potentials to reproduce desirable ion concentrations, therefore several trial runs were necessary to fit the chemical potential. At the same concentrations, the NVT and mVT simulations reproduce identical results.
The fraction of bound ions, Pi, of species i is determined as a ratio of the number of "bound", or "associated" ions to the total number of these ions in the solution. The ions are considered as bound if the distance between the ion and the polyion surface is less than some value rb, which is often taken to be 5 Å 31,44, the typical radius of the short-range van der Waals interactions. The number of bound ions are then obtained by integrating the radial density distribution between the polyion surface and this value rb. It was shown in previous work45 that within reasonable limits, the variation of the value of rb in the case of simple ions was of minor importance in the calculation of Pi. A problems arises how to count bound polyamines if only a part of a molecule occurs within the distance rb. From one point of view we can consider this molecule as "partially bound" (e.g., if only one of the three monomers of our spermidine model occurs within the distance rb from the DNA surface, this gives a contribution 1/3 to the total number of bound ions). This definition enables Pi to be calculated by integrating the radial distribution function, as in the case of simple ions. Another definition may be that the whole spermidine molecule is considered as bound already if only one of its monomers occurs within the distance rb (we denote the second definition as P*i). Comparison of the two definitions is given in Fig.1 for different values of rb for the case of 20 mM, 200 mM Na+ and 120 mM DNA solution. One can see that the second definition yields slightly higher and more stable (less dependent on rb) values of the fraction of bound polyamine ions at the same rb. Below we will give both values, Pi for 5 Å and Pi* for rb=3 Å. In the case of the grooved DNA model the distance rb is counted out from a fictitious surface at 10 Å from the DNA axis.
4. Results and Discussion
A. Ion distributions. Case of monovalent salt.
The radial density profiles of the three-valent ions for the four considered models are displayed in Fig. 2 and the corresponding integral charge functions I(r) are shown in Fig. 3:
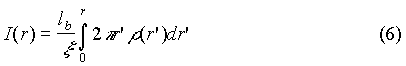
This function also defines the fraction of bound ions by P=I(rb)cp/ci where cp and ci are the phosphate and mean ion concentrations respectively.
The concentration of three-valent ions was 0.02 M, the concentration of Na+ in supporting 1:1 salt was 0.2 M and the DNA concentration was 0.12 M, corresponding to a DNA-DNA distance of 97Å. Here and below we consider "concentration" as "mean" concentration, defined by the total number of ions in the cell divided by the volume: ci=cmean=Ni/V .
Comparison of the PB and SI models confirms the well known picture that the Poisson-Boltzmann equation underestimates the density of the most highly charged counterions in the close vicinity of the polyion. The Poisson-Boltzmann theory neglects ion-ion correlations, which produce additional attractive contributions and increase the ion density close to the polyion surface compared to Eq. (1). It should be noted that the SI curve exceeds the PB curve for small distances (r<12Å), which is barely seen in the figure because of the logarithmic scale of the Y axis and the very steep slope of the curves (see inset in Fig. 2). The higher spermidine counterion concentration in the SI model is more clearly seen in Fig. 3, where the integrated charge, I(r) is displayed. In the SI model more counterions are accumulated within a small (2 Å thickness) layer near the polyion surface, and hence the concentration at other distances (in the bulk solution) is essentially lower than in the PB theory, since the total number of ions is equal in both cases.
Substitution of the spherical 3-valent ions by the 3-valent polyamine chain (HC model) leads to an opposite effect. The counterion density close to the DNA surface is even lower than what follows from the PB theory. Several reasons for this behavior may be discerned. The interaction between a polyvalent counterion with three distributed charges and a polyelectrolyte surface, is weaker than that of a three-valent point charge counterion. In fact, at short distances from the DNA polyelectrolyte surface, the interaction is more like that of monovalent counterions. Furthermore, the correlations between two distributed charges are weaker than it is between two point charges. In addition, the spermidine counterions in the HC and GD models have internal degrees of freedom and a corresponding entropy contribution to the free energy. A spermidine molecule bound to a polyion has less space to move in, and hence, lower entropy as compared to the same molecule in the bulk. This creates an effective repulsive force for the polymeric counterions. The entropy contribution to the binding of flexible ionic ligands to polyelectrolytes was recently calculated also in ref.33 with a lattice model in combination with the PB theory. This contribution was found to decrease the binding coefficients for flexible ligands essentially, as in the case of the present study. It should however be noted the predictions of the this PB-lattice model is still expected to overestimate the binding since the electrostatic contribution to the binding constant is still calculated within a point charge PB description. Exact comparison of the results between these two approaches is, however, difficult without additional calculations since the systems and experimental conditions modeled differ considerably.
The grooved (GD) model takes into account the specific charge distribution and some of the aspects of the real grooved structure of the DNA surface. The counterions can penetrate in the grooves, giving a non-zero density distribution at distances r<10 Å. In general, however, the cylindrically averaged density distribution in the vicinity of the polyion is lower compared to the case of a hard cylinder. For large distances from the polyion the cylindrical and the grooved models give very similar density distributions and integral charge curves. The effects of a helical charge distribution with a grooved DNA surface have been studied in previous work19,20,46 for monovalent ions and a similar picture was observed as in the present work for spermidine counterions. The density profile for the grooved model in Fig. 2 is a cylindrically averaged density. The real three-dimensional ion density in a plane perpendicular to the DNA axis is shown in Fig. 4. The maximum counterion density occurs in the minor DNA groove, and it reaches a value around 1 M, just as in the case of a hard cylindrical polyion. For distances greater than 5 Å from the surface, the ion density has an almost perfect cylindrical symmetry. One should, however, note that our model includes only the electrostatic interactions due to the ionic nature of the system. Taking into account specific ion-DNA interactions may redistribute the counterion density.
The monovalent ion densities for the considered cases are shown in Fig. 5. For the monovalent counterions one can note an opposite effect as compared to the spermidine density distribution. The ion density in the vicinity of the polyion is lower for the SI model and higher for the HC model, compared to the PB theory. For monovalent ions the PB mean field theory is almost applicable, and the density distribution generally satisfies Eq. (1). A higher concentration of the three-valent ions close to the polyion means lower electrostatic potential and consequently lower monovalent ion density.
Comparison of the ion density for the models PB and HC at different 1:1 salt concentrations is given in Fig. 6. Addition of monovalent salt leads to displacement of three-valent ions from a region near DNA to bulk solution. The difference in results between the models is about the same at the different salt concentrations.
B. Spermidine ion distributions. Effects of competition by divalent salt.
The spermidine, magnesium and chloride ion density distributions in solutions with addition of 2:1 (e.g. MgCl2) salt are given in Fig. 7. The spermidine distributions show differences due to effects of competition with the two kinds of added salt, NaCl and MgCl2 respectively. Generally, the relationships between the results corresponding to the different models are qualitatively similar to the case of monovalent salt, but quantitatively the differences between the models are more pronounced when magnesium is the competitive counterion. The spermidine concentration near the DNA polyelectrolyte surface is lower for Mg2+ as the competing counterion. This effect is, however, not very pronounced for the PB model with spermidine and Mg2+ treated as three- and two-valent point charges. Thus, within the much more realistic MC models treating spermidine as a polyvalent molecule with distributed charges, the magnesium counterions are able to compete more effectively for association to DNA. Regarding the magnesium ion density distribution, as for the case of the sodium distributions, the concentration close to the DNA surface is higher within the HC model than it is for the PB model.
In the presence of divalent ions, the counterion cloud may not only compensate the polyion charge, but even exceed it, resulting in opposite values of the electrostatic potential at some distances. This effect, called "overneutralisation", or "charge reversal", can not appear in the Poisson-Boltzmann theory, but has been observed in other theoretical studies: Monte-Carlo simulations12 and HNC calculations8 for polyelectrolyte systems with divalent counterions and even for monovalent counterions at high concentrations20. Does the charge reversal occur in real systems or it is an artifact of the model? We are aware of one rather old experimental evidence for this phenomenon by Strauss et al47, who observed that a cationic polyion (polysoap) during electrophoresis moved against the electric field at high salt concentrations. The underlying reason of the charge reversal is attractive forces arising from the correlation of ions; the same effect is responsible for effective attraction between likely charged polyelectrolytes which occurs is some cases19 and leads e.g. to precipitation or condensation of DNA into a compact and ordered phase48.
Results of the electrostatic potential for the different models and under different conditions are given in Fig. 8. One can note the following: An addition of both divalent and spermidine counterions makes the charge reversal stronger. At low concentrations the effect does not exists at all. Taking the detailed structure of the DNA surface into account (GD model) leads to a weaker effect, but it does not disappear.
C. Binding properties
In the following analysis we concentrate on global properties of the ion distributions and will present results of calculations performed under varying solution conditions for the different models. The properties that have been calculated are the fraction of bound ions, Pi, the ratio of the mean ion concentration Cmean=Ni/V to the concentration at the outer cell boundary R, x=cmean/cbulk, and the electrostatic potential at the surface of the polyion. All these quantities are determined by the ion distribution as a whole, and they are interconnected with each other. They generally characterize the balance between those ions associated with the polyion and those remaining in the bulk and are important quantities in interpretations of many experimental results as for example in counterion NMR experiments24,29,31,44. Values of Pi define the so called "localized binding"24,33, which refers only to ions in close proximity to the polyion. Values of x are related to the "excess" ligand concentration cmean-cbulk=cbulk(x-1) which is also directly related to the Gibbs definition of binding33. Values of x define in fact the "delocalized" binding which is measured in a Donnan membrane equilibrium, if it is assumed that the ion concentration at the outer cell boundary, c(R), is equal to the bulk concentration in a Donnan membrane equilibrium. This will be correct if the electrostatic potential is zero and all ion densities are constant near the outer cell boundary, which is usually the case if the concentration of coions exceeds the concentration of charged groups of the polyions. The fraction of "delocalized bound ions" is expressed as Pd=(cmean-cbulk)/cbulk=1-1/x. This number does not depend on the specific definition of the radius rb, and it include also ions which may be far from the polyion.
The dependencies of the fraction of bound ions on the monovalent salt concentration for different models are given in Table 1, and results for the ratio x=cmean/cbulk and for the surface electrostatic potential are given in Table 2. It can be seen that the spherical three-valent ions (the SI model) are attracted to DNA more strongly than what follows from the PB theory. At low salt concentrations almost all three-valent ions occur in the vicinity of DNA and there are practically no such ions in the bulk. However, taking into account the internal structure of the spermidine molecule reduces the amount of bound ions to values even lower than the PB result. Additional refinement of the model (the grooved model) leads to a further decrease of the fraction of bound ions, especially at high monovalent salt concentration. For monovalent counterions we have an opposite effect: in all simulations Pi for monovalent ions occur higher compared to the PB results. This difference in the results is of course qualitatively similar to the trends that were observed for the ion distributions. It shows that two main approximations within the present mean field PB description of polyamine-DNA interactions, the neglect of ion correlations and the model of spermidine as a three-valent point charge, to some extent cancel each other. The results also show that including the structural features of the spermidine polyamine, particularly in combination with the helical charge distribution on the DNA, has an important effect. From the results of Tables 1 and 2 it can thus be concluded that the model describing the spermidine molecule as a spherical three-valent ion is not suitable for modeling the ion binding properties of this polyamine.
There exists another simple and widely used theory of ion binding competition based on Mannings condensation theory, see e.g. ref.16. This theory is, however, not directly compatible with our simulations mainly because it implies a single polyion, or at least a very low polyion concentration compared to the ion concentrations, which is not the case in the present work. A formal application of the expressions of reference16 to the parameters in Tables 1 and 2 yields considerably higher binding constants than the present refined (grooved) model, more close to the results of the SI model.
Experimental NMR studies of the self-diffusion coefficients of methylspermidine in DNA solutions under different salt conditions that have been performed in our laboratory, confirm that the Poisson-Boltzmann theory overestimates the amount of polyamine molecules in the close vicinity of DNA24. Within the two-state self-diffusion model, the retardation of the counterion self-diffusion coefficient, D, is directly related to the fraction of bound counterions, Pi. It was found in ref.24 that experimental values of D are higher than the theoretical ones calculated on the basis of the PB theory within this two state model. This implies experimentally a smaller Pi and less strong polyamine association, than what is predicted by the PB theory. Furthermore, it was observed that the difference was larger at higher NaCl concentrations or in the presence of the divalent Mg2+ ion. Our simulations, although carried out for other ranges of concentration, also shows that a chain of three monovalent ions is a weaker competitor than a simple three-valent ion, even if the three-valent ions are treated within the Poisson-Boltzmann approximation. However, a chain of three monovalent ions is more strongly attracted to the DNA polyion surface than monovalent ions are, since the calculated coefficients PSpd are always higher than Pi for monovalent ions. It is also of interest to note that the NaCl salt dependence of PSpd in Table 1 is stronger in the more realistic models than the result obtain within the PB treatment. This is in accordance with the NMR results of ref.24, where the polyamine diffusion was found to be considerably more salt dependent than what could be predicted from a PB treatment.
A comparison of the grooved model with the PB theory at different spermidine concentrations is given in Table 3. Once again one can see that the PB theory generally overestimates the binding of spermidine ions to DNA with the exception of high spermidine concentrations. In addition, it can be noted that for small amounts of added spermidine (less than 0.01M), the effect on PSpd due to increasing its concentration is larger in the more refined models, as compared to the PB treatment. This is also consistent with the experimental observations at higher amounts of added NaCl salt. Those experimental data, interpreted within the two state model show that PSpd varies more with the spermidine concentration that the prediction of the PB model.
Results for a system consisting of DNA, spermidine molecules, divalent counterions (e.g., Mg2+) and monovalent coions are given in Tables 4, 5 and 6. As in the case of monovalent salt, the PB theory generally overestimates the binding of spermidine to DNA as compared to the MC results with a discrete charge distribution on the spermidine molecules, the difference being greater for small spermidine concentration and for high Mg2+ concentration. The simulation results also show that the binding of Mg2+ ions to DNA occurs noticeably stronger as compared to the PB theory.
An interesting observation concerns the relationship between the cylindrical and the grooved model for DNA in the presence of Mg2+. Contrary to the case of monovalent salt, PSpd for the grooved model is smaller than PSpd for the cylindrical model only at low salt concentrations. At high Mg2+ concentration the grooved model has a larger spermidine binding than the primitive model. This may be related to the effect of charge reversal which creates a repulsive contribution to the force for spermidine molecules at distances 5-10 Å from the surface. The effect of charge reversal is just stronger for the primitive model and for high Mg2+ concentrations.
Another observation is that the fraction of bound ions is about the same for spermidine and for Mg2+ for the refined MC models. This means that the Mg2+ and Spermidine3+ ions compete about equally for association to the DNA surface. There is also what seems to be a screening effect at higher concentrations of total added multivalent salt, where Mg2+ occurs to be a somewhat more effective competitor (PMg>PSpd), whereas at low total multivalent salt concentrations there is an opposite relation: PSpd>PMg. The competitive behavior of Spermidine3+ therefore appears to be reduced from that expected for a three-valent point charge, to such an extent that it is nearly equal to that of a two-valent point charge. This observation is in qualitative agreement with equilibrium dialysis experiments, where similar binding constants were obtained for the NaCl salt dependence of Mg2+ binding to poly(rArU) as for Spermidine3+ binding to calf thymus DNA49,50. Indirect evidence for this similarity was also obtained from the observation that Mg2+ binding to calf thymus DNA in the presence of Spermidine3+ is intrinsically stronger than that of the two valent polyamine Putrescine2+,50. Furthermore, 23Na NMR measurements gave similar values of n0, the number of associated Na+ ions displaced for each multivalent ion added, upon titration with Mg2+ or Spermidine3+ to calf thymus DNA29. This was in contrast to the results upon titration with Putrescine2+ and Co(NH3)63+, where the n0 values were smaller and larger respectively.
Table 6 show dependencies of the binding properties on the spermidine concentration for two cases: a low Mg2+ concentration corresponding to DNA charge compensation (A) and a higher Mg2+ concentration (B). It can be seen that PSpd behaves differently; it changes with Spermidine concentration in the case A and preserves almost the same value in the case B. The PB theory predicts essential changes of PSpd in both cases. The case A is clearly the most relevant for comparison with the NMR experiments where diffusion of Spermidine was measured as a function of this polyamine concentration in MgDNA solutions24. It can again be noted that the experimental observation within the two state model, is that PSpd varies more with the Spermidine concentration than the prediction of the PB model.
5. Conclusions
In this work we have studied several models of a DNA polyelectrolyte system containing a mixture of simple and complex multivalent ions like the polyamine spermidine, within the frame of a continuum dielectric approach. We have shown that the behavior of complex multivalent ions with distributed charges differs strongly from the properties of atomic point charge ions of the same valence. The Poisson-Boltzmann theory can be considered only as an approximation for this system. Agreement between some results from the PB theory and the most refined MC simulations that occur under certain conditions is mainly due to cancellation of different factors introduced by the approximations within the PB model. It is clear that computer simulations provide a more solid basis for calculations of different properties of the ion distributions, yielding a statistically correct description within the frame of the chosen model. Further refinement of the present models may include some account for the molecular structure of the solvent and for specific short-range ion-DNA interactions. This may be done on the basis of effective ion-ion and ion-DNA interactions40. Investigations along these lines are presently in progress in this laboratory.
Comparison of the simulation results with experimental NMR diffusion data studying the interaction of the methylated spermidine polyamine analogue with DNA24 is interesting since it corroborates a picture of the polyamine-DNA association as governed largely by electrostatic interactions. From this comparison it is also clear that the MC simulations within a description of the polyamine treated with three distributed discrete charges (HC or GD model), gives a qualitatively better description of the effects of polyamine, NaCl and MgCl2 concentrations on the polyamine association to DNA, than does the PB treatment. Experimental support for these observations is also found in studies of polyamine - nucleic acid binding, using equilibrium dialysis49,50 and 23Na NMR29.
Acknowledgments.
This work has been supported by the Swedish Natural Science Research Council (NFR), the Wenner Gren Foundations and by the Swedish Royal Academy of Sciences (KVA).
References and Notes
(1) Manning, G. S. Quart. Rev. Biophys. 1978, 11, 179.
(2) Record, M. T.; Mazur, S. J.; Melancon, P.; Roe, J. H.; Shaner, S.; Unger, L. Annu. Rev. Biochem., 1981, 50, 997.
(3) Sharp, K.; Honig, B. Annu. Rev. Biophys. Biophys. Chem., 1990, 19, 301.
(4) Manning,G.C. J.Chem.Phys, 1969, 51, 924.
(5) Fuoss, R. M.; Katchalsky, A.; Lifson, S. Proc. Natl. Acad. Sci.USA, 1951, 37, 579.
(6) Fixman, M,; J. Chem. Phys., 1979, 70, 4995.
(7) Bacquet, R. J.; Rossky, P. J. J. Phys. Chem., 1984, 88,2660.
(8) Tovar, E. G.; Losada-Cassou, H.; Henderson, D. J. Chem. Phys., 1985, 83, 361.
(9) LeBret, M., Zimm, B. Biopolymers, 1984, 23, 271.
(10) Murthy, C. S.; Bacquet, R. J.; Rossky, P. J. J. Phys. Chem 1985, 89, 701.
(11) Mills, P.; Anderson, C. F.; Record, M. T. J. Phys. Chem. 1985, 89, 3984.
(12) Vlachy, V.; Haymet, A. D. J. J. Chem. Phys. 1986, 84, 5874.
(13) Jayaram, B.; Beveridge, D. L. J. Phys. Chem. 1991, 95, 2506.
(14) Guldbrand, L.; Nilsson, L. G.; Nordenskiöld, L. J. Chem. Phys. 1986, 85, 6686.
(15) Vorontsov-Velyaminov, P. N.; Lyubartsev, A. P. Molec. Simulations 1992, 9,285.
(16) Wilson, R. W.; Bloomfield, V. A. Biochemistry 1979, 18, 2192.
(17) Rouzina, I., Bloomfield, V. A. J. Phys. Chem. 1996, 100, 4292.
(18) Klein, B. J.; Pack, G. R. Biopolymers 1983, 22, 2331.
(19) Lyubartsev, A. P.; Nordenskiöld, L. J. Phys. Chem., 1995, 99, 10373.
(20) Montoro, J. C. G.; Abascal, J. L. F. J. Chem. Phys., 1995, 103, 8273.
(21) Fenley, M. O.; Manning, G. S.; Olson, W. K. J. Phys. Chem. 1992, 96, 3963.
(22) Anderson, C. F.; Record, M. T., Jr. Annu. Rev. Phys. Chem. 1982, 33, 191.
(23) Frank-Kamenetskii, M. D.; Anshelevich, V. V.; Lukashin, A. L. Sov. Phys. Uspekhi, 1987, 30, 317.
(24) Andreasson, B; Nordenskiöld, L.; Schultz, J. Biophys.J. 1996, 70,2847.
(25) Saenger,W. in Priciples of Nucleic Acid Structure, Springer-Verlag: New York, 1984.
(26) Drew, H. R.; Dickerson, R. E. J.Mol.Biol. 1981, 163, 129.
(27) Jain, S.; Zon, G.; Sundralingam, M. Biochemistry, 1989, 28, 2360.
(28) Burton, D. R.; Forsen, S.; Reimarsson, P. Nucleic Acids Res., 1981, 9, 1219.
(29) Braunlin, W. H.; Anderson, C. F.; Record, M. T. Biopolymers, 1986, 25, 205.
(30) Plum, E. G., Bloomfield, V. A. Biopolymers, 1990, 29,13.
(31) Andreasson, B.; Nordenskiöld, L., Braunlin, W. H.; Schultz, J., Stilbs, P. Biochemistry, 1993, 32, 961.
(32) Rouzina, I.; Bloomfield, V. A. J.Phys.Chem., 1996, 100, 4305.
(33) Stigter, D.; Dill, K. A. Biophys. J. 1996, 71, 2064.
(34) Wennerström, H.; Jönsson, B.; Linse, P. J.Chem.Phys., 1982, 76, 4665.
(35) Reiner, E. S.; Radke, C. J. Adv.Colloid and Interface Sci., 1993, 47, 59.
(36) Hecht, J. L.; Honing, B.; Shin, Y. K.; Hubbel, W. L. J.Phys.Chem., 1995, 99, 7782.
(37) Hansen, W. N. J. Electroanal. Chem., 1983, 150,133.
(38) Pratt, L. R.; Hummer, G.; Garcia, A. E. Biophys. Chem., 1994, 51,147.
(39) Guardia, E.; Rey, R.; Padro, J. A. J. Chem. Phys., 1991, 95, 2823.
(40) Lyubartsev, A. P.; Laaksonen, A. Phys. Rev. E., 1995, 52,3730.
(41) Rashin, A. A.; Bukatin ,M. A. Biophys. Chem., 1994, 51, 167.
(42) Triolo, R.; Grigera, R.; Blum, L. J. Phys. Chem., 1976, 80, 1858 .
(43) Corti, H. R.; Fernandez-Prini, R. J.Chem.Soc., Faraday Trans.2, 1986, 82, 921.
(44) Braunlin, W. H.; Drakenberg, T.; Nordenskiöld, L. Biopolymers, 1987, 26, 1047.
(45) Bleam, M. L.; Anderson, C. F.; Record, M. T. Biochemistry, 1983, 22, 5418.
(46) Mills, P.; Paulsen, M. D.; Anderson, C. F.; Record, M. T. Chem.Phys.Lett., 1986, 129, 155.
(47) Strauss, U. P.; Gershfeld, N. L.; Spiera, H. J. Am. Chem. Soc. 1954, 76, 5909.
(48) Leikin, S.; Parsegian, V. A.; Rau, D. C.; Rand , R. P. Annu. Rev. Phys. Chem, 1993, 44, 369.
(49) Krakauer, H. Biopolymers, 1971, 10, 2459.
(50) Braunlin, W. H.; Strick, T. J.; Record, M. T. Biopolymers, 1982, 21, 1301.
Table 1.
Binding properties of ions in different models of Spermidine-NaCl-DNA solution. Effect of the monovalent salt concentration. [DNA-P]=0.12 M, [Spd]=0.02 M. Binding radius 5 Å for P and 3 Å for P* (see Sec.3)
---------------------------------------------------------------------------
PB Spherical ions Cylindrical model Grooved model
[Na+] P+3 P+1 P+3 P+1 Pspd P*spd P+ Pspd P*spd P+
--------------------------------------------------------------------------
0.1 0.94 0.26 0.995 0.31 0.86 0.91 0.33 0.85 0.90 0.32
0.2 0.87 0.19 0.97 0.21 0.76 0.81 0.23 0.75 0.79 0.22
0.5 0.67 0.13 0.83 0.14 0.52 0.58 0.16 0.41 0.45 0.15
1. 0.47 0.10 0.66 0.08 0.30 0.34 0.11 0.23 0.26 0.12
---------------------------------------------------------------------------
Table 2.
Ratio of mean to bulk concentrations x=cmean/cbulk and the surface electrostatic potential in different models of Spermidine-NaCl-DNA solution. Effect of the monovalent salt concentration. Concentrations: [DNA-P]=0.12 M, [Spd]=0.02 M
-------------------------------------------------------------------------
x(+3 ions) x(+1 ions) surface potential -ey/kT
C(Na+) PB SI CP GD PB SI CP GD PB SI CP GD
-------------------------------------------------------------------------
0.1 57 inf 50 45 1.47 1.67 1.69 1.72 3.92 2.55 3.05 3.11
0.2 13 500 14 13 1.2 1.27 1.32 1.3 2.63 1.91 2.43 2.57
0.5 3.1 5.7 3.2 2.8 1.05 1.07 1.08 1.14 2.02 1.41 1.82 2.11
1 1.8 3.3 1.9 1.7 1.03 1.05 1.03 1.03 1.71 0.95 1.36 1.62
-------------------------------------------------------------------------
Table 3.
Effect of spermidine concentration on the fraction of bound ions P, on the ratio of mean to bulk concentrations and on the surface electrostatic potential. A comparison of the grooved model of DNA (GD) and the PB approximation. Concentrations: [DNA-P]=0.12 M, [Na+]=0.2 M
-----------------------------------------------------------------------
Spd Na x(Spd) x(Na) potential
GD *GD PB GD PB GD PB GD PB GD PB
------------------------------------------------------------------------
0.004 0.85 0.89 0.98 0.32 0.31 80 130 1.69 1.64 2.94 3.49
0.01 0.80 0.85 0.96 0.27 0.25 40 52 1.52 1.43 2.85 3.12
0.015 0.76 0.81 0.92 0.26 0.21 22 20 1.37 1.29 2.72 2.83
0.02 0.73 0.78 0.87 0.22 0.19 16 13 1.3 1.2 2.57 2.64
0.05 0.49 0.53 0.55 0.14 0.12 2.7 2.6 1.08 1.06 1.87 1.8
0.1 0.29 0.32 0.33 0.11 0.09 1.7 1.4 1.02 1.03 1.67 1.45
-----------------------------------------------------------------------
Table 4.
Fraction of bound ions in different models of Spermidine-MgCl-DNA solution. Effect of the divalent ions concentration. Concentrations: [DNA-P]= 0.12 M, [Spd]=0.02 M
------------------------------------------------------------------
PB Cylindrical model Grooved model
c(Mg) P+3 P+2 Pspd P*spd Pmg Pspd P*spd Pmg
-------------------------------------------------------------------
0.04 0.84 0.51 0.74 0.83 0.73 0.70 0.76 0.70
0.1 0.63 0.31 0.42 0.48 0.45 0.38 0.41 0.43
0.2 0.44 0.22 0.22 0.25 0.29 0.22 0.25 0.27
0.5 0.23 0.13 0.09 0.10 0.16 0.11 0.12 0.15
-------------------------------------------------------------------
Table 5.
Ratio of mean to bulk concentrations x=cmean/cbulk and the surface electrostatic potential in different models of Spermidine-Mg-Cl-DNA solution. Effect of the divalent ion concentration. Concentrations: [DNA-P]=0.12 M, [Spd]=0.02 M
----------------------------------------------------------------------
x(+3 ions) x(+2 ions) surface potential
[Mg2+] PB CP GD PB CP GD PB CP GD
----------------------------------------------------------------------
0.04 11.8 13 16 2.74 4.7 4.5 2.52 1.69 1.88
0.1 2.9 2.0 1.95 1.47 1.7 1.75 1.95 1.16 1.46
0.2 1.7 1.21 1.28 1.2 1.32 1.28 1.64 0.86 1.18
0.5 1.2 1.0 1.02 1.05 1.08 1.08 1.28 0.50 0.86
-----------------------------------------------------------------------
Table 6.
Effect of spermidine concentration on the fraction of bound ions P, ratio of mean to bulk concentrations and on the surface electrostatic potential. A comparison of the grooved model and the PB approximation.
------------------------------------------------------------------------------------
x=c(mean)/c(bulk)
Pspd P(Mg) x(spd) x(Mg) potential
c(Spd) GD *GD PB GD PB GD PB GD PB GD PB
-------------------------------------------------------------------------------------
A) [DNA-P]=0.05M, [Mg+2]=0.025M
0.002 0.75 0.80 0.92 0.71 0.60 25 50 6 4.5 2.37 3.33
0.005 0.63 0.67 0.87 0.62 0.47 6.6 16 3.6 2.7 2.15 2.94
0.01 0.55 0.59 0.74 0.49 0.33 3.9 5.6 2.3 1.8 1.97 2.51
0.02 0.40 0.43 0.51 0.37 0.20 2.0 2.3 1.67 1.32 1.71 2.08
0.05 0.21 0.23 0.26 0.25 0.11 1.43 1.36 1.24 1.11 1.55 1.65
B) [DNA-P]=0.12M, [Mg+2]=0.2M
0.004 0.23 0.25 0.53 0.29 0.25 1.37 2.0 1.34 1.27 1.21 1.78
0.01 0.23 0.25 0.5 0.28 0.24 1.31 1.9 1.31 1.24 1.17 1.72
0.02 0.22 0.24 0.44 0.27 0.22 1.28 1.7 1.28 1.2 0.89 1.64
0.05 0.21 0.23 0.34 0.24 0.17 1.25 1.43 1.21 1.12 0.85 1.47
0.1 0.17 0.19 0.24 0.2 0.14 1.2 1.23 1.15 1.07 0.81 1.29
0.2 0.14 0.16 0.17 0.15 0.11 1.14 1.11 1.08 1.03 0.78 1.11
------------------------------------------------------------------------------------>
Figures.
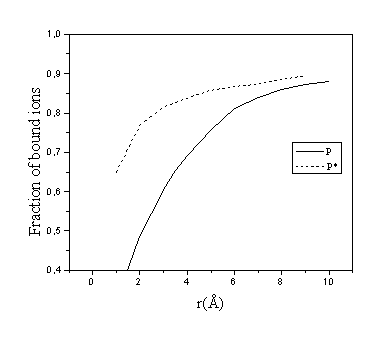
Fig. 1. Dependence of two alternative definitions of fraction of bound spermidine ions on the choice of cutoff distance rb. Concentrations: [DNA-P]=0.12 M, [Spd]=0.02 M, [Na+]=0.2 M
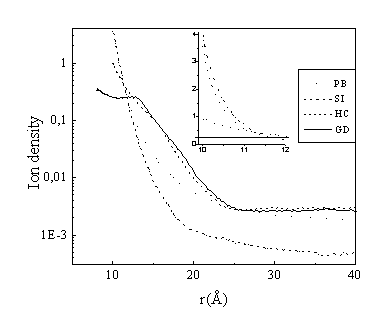
Fig. 2. Density distribution of spermidine ions in different models. The inset shows the behaviour close to DNA with linear scale of the Y axis. Concentrations:
[DNA-P]=0.12 M, [Spd]=0.02 M, [Na+]=0.2 M
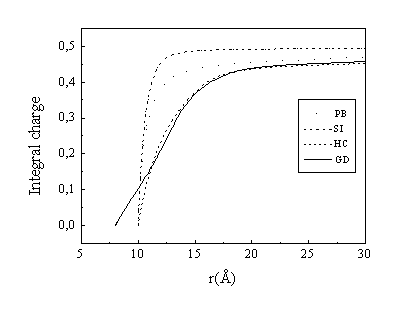
Fig. 3. Integral charge I(r) of spermidine molecules in different models. Concentrations:
[DNA-P]=0.12 M, [Spd]=0.02 M, [Na+]=0.2 M
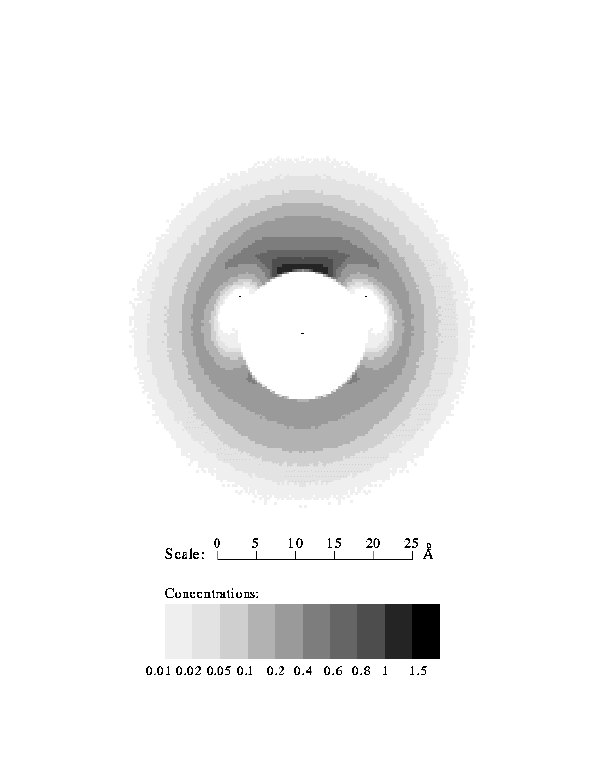
Fig. 4. Monomer density of spermidine molecules in the plane perpendicular to the DNA axis. Grooved model of DNA.
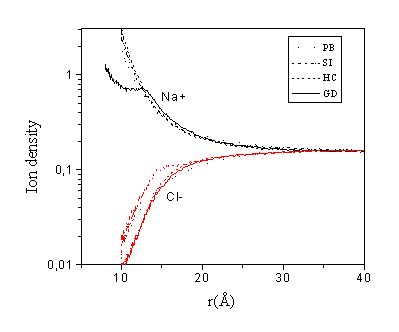
Fig. 5. Density distribution of monovalent ions in different models. Concentrations:
[DNA-P]=0.12 M, [Spd]=0.02 M, [Na+]=0.2 M
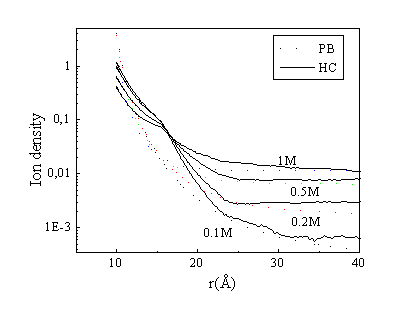
Fig. 6. Density distribution of spermidine molecules at different monovalent salt concentrations. A comparison of the PB theory and the hard cylinder (HC) model. Concentrations: [DNA-P]= 0.12 M, [Spd]=0.02 M.
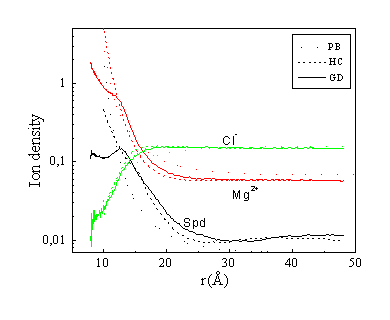
Fig. 7. Density distribution of Spd3+, Mg2+ and Cl- ions in different models. Concentrations:
[DNA-P]=0.12 M, [Spd]=0.02 M, [Mg2+]=0.1 M
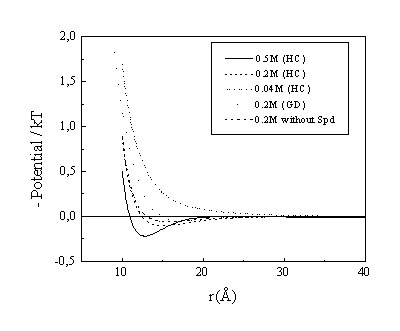
Fig. 8. Electrostatic potential around DNA in mixture of spermidine, Mg2+ and Cl- ions. Mg2+ concentration is on the figure, [Spd]=0.02 M (except the last line), [DNA-P]=0.12 M.