NFORM = <format>
where <format> is one of:
MDYN- MDynaMix binary trajectory (default)XMOL- XMOL trajectory. It is implied, that the commentary (second) line of each configuration is written in the format:(char) <time> (char-s) BOX: <box_x> <box_y> <box_z>
where
(char)is any character word,<time>is time in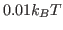 ,
,
<box_x> <box_y> <box_z>(following after keywordBOX) are the box sizes.PDBT- PDB trajectory as generated by ``trajconv'' utility of GROMACS simulation package.DCDT- DCD trajectories generated by NAMD package
DCD_timefactor = <value>
Time scaling factor in DCD trajectories. Default: 0.048888 ps
FNAME = <file_name>
set the base name of the trajectory files. The trajectory must be written
as a sequence of files <file_name>.001 , <file_name>.002 and
so on, the largest possible number being <file_name>.9999 .
PATHDB = <value>
Directory with molecular description files (.mmol). Default is the current directory (.) .
NTYPES = <value>
Number of molecule types in the trajectory
NAMOL = <name1> [,<name2>,...]
NTYPES names of molecules. It is supposed that files
<name1>.mmol, <name2>.mmol,... describing the molecules are
present in the directory defined by PATHDB. Format of .mmol files is
the same as for MDynaMix program. For analyzing trajectories generated by
other programs, .mmol files are still used to provide information about atomic
masses and charges. It is however enough to have only the first section of .mmol files
containing description of atoms.
The program may work without .mmol files, if parameters NSPEC and
NSITS (see below) are given. In this case, the masses of all atoms are set to
1 and the charges to 0, which will result in definition of CG sites as geometric centers of
the atomic groups, and zero charges of CG sites (the later can be manually corrected at
the next stage).
NSPEC = <n1>[,<n2>,...]
Number of molecules of each type ( NTYPES numbers). This parameter
is not necessary in MDynaMix binary trajectories.
NSITS = <n1>[,<n2>,...]
Number of atoms on each molecule of each type ( NTYPES numbers).
This parameter is not necessary if .mmol files for each molecular type are
provided.
NFBEG = <value>
Number of the first trajectory file (integer between 0 and 9999)
NFEND = <value>
Number of the last trajectory file (integer between 0 and 9999)
TSTART = < value>
Start reading the trajectory file(s) after configuration with this value of time (in ps). Default: 0.
TOFFSET = <lavue>
While writing the CG trajectory, add TOFFSET ps to each time stamp of the atomistic trajectory
IPRINT = <value>
Defines how much you see in the intermediate output. The final output with analysis of results does not depend on it. Default value is 5.
BOXL = <x-box-size>
BOYL = <y-box-size>
BOZL = <z-box-size>
define the box size if it is not present in the trajectory (implies constant-volume simulation) If information of box sizes is present in the trajectory, box size parameters from the input file are ignored.
ISTEP = <value>
Specifies that only each ISTEP-th configuration from the trajectory
is taken for the analysis. D§efault is 1.
The second part of the main input file for cgtraj utility, which describes CG bead
mapping scheme, has a hypertext-like format.
The section starts with keyword: BeadMapping and ends with EndBeadMapping.
Each coarse grain molecular type is described in a separate section, which starts with tag
CGMolecularType: <CGMolecularTypeName> and ends with EndCGMolecularType.
Inside each section, parental molecular type name and CG beads definition should be given.
The parent's name is defined by the tag
ParentType: <ParentMolecularTypeName>.
CG beads are defined in a line-per-bead way, where every line has the following structure:
<Bead name>:<Number of atoms in the bead>:<list of atoms atom1,atom2,...>,
where list of atoms is a comma separated list of atom numbers according to the mmol-file
describing parental molecular type. A file named <CGMolecularTypeName>.CG.mmol
containing description of a coarse-grained molecule will be created for every defined
CG-molecular type.
The keywords/tags are not case sensitive, and spaces will be automatically removed from the text.